Back to Golden Gate Protocols
Part Plasmid Assembly
Once you have designed your part and either amplified with PCR or ordered the desired gBlock (as described
here
) you can proceed to the assembly step of the part plasmid itself. The example reaction below shows pYTK001 used as the entry vector for the reaction; however, this can be substituted for any other entry vector with requisite
BsmBI cut sites. Once built, part plasmids can be assembled into transcriptional units.
Protocol source: NEB (
https://www.neb.com/protocols/2020/01/15/golden-gate-assembly-protocol-for-using-neb-golden-gate-assembly-kit-bsmbi-v2-neb-e1602
)
Assembly Reaction
Access the old non-kit golden gate assembly protocols here
1. Calculate the mass (in ng) required for 50 fmol of vector and 100 fmol of insert using NEB's
NEBioCalculator:
https://nebiocalculator.neb.com/#!/dsdnaamt 2.
2. Set up the following reaction mix:
- 50 fmol pYTK-001 plasmid = 82.68 ng [try to keep volume to 1-2μL]
- 100 fmol of insert DNA = 650 * insert length * 100x10^-6 = X ng [try to keep volume to less than 10 μL]
- 2 μL of NEB 10× T4 DNA ligase buffer
- 1 μL of NEB Golden Gate Enzyme Mix (BsmBI-v2)
- x μL water up to 20 μL total
3. Mix samples well by pipetting, then run the reaction on the thermocycler under the following conditions:
Optionally, you may have high enough efficiency if inserting one PCR product into the entry vector with NEB's short protocol with no cycling instead of the longer protocol:
4. Transform 2 μL assembly reaction into
Electrocompetent or
Chemically Competent cells and plate on LB + Cam
Expected Results
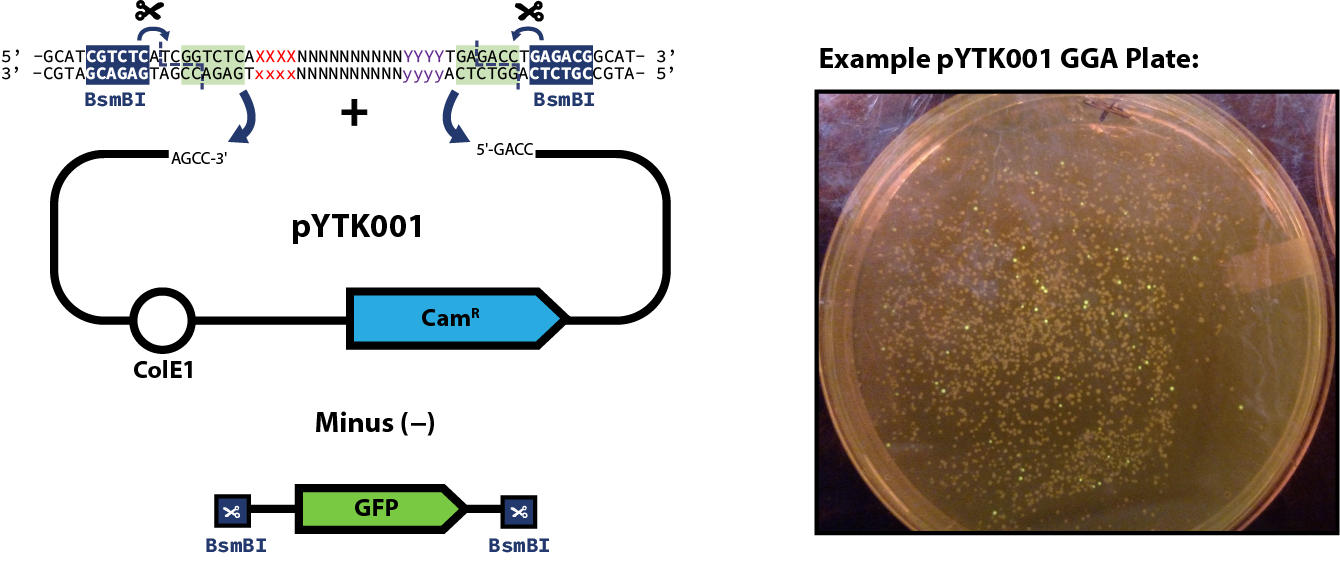
- (If using pYTK001 as your entry vector) Multiple non-fluorescent colonies
- Be sure to screen colonies for GFP expression on a blue light transilluminator, pick colonies that do not fluoresce!
- Cells containing properly assembled plasmids (confirmed through PCR and Sanger Sequencing)
If you are having repeated issues check out the Troubleshooting page!
Back to Golden Gate Protocols
 ) you can proceed to the assembly step of the part plasmid itself. The example reaction below shows pYTK001 used as the entry vector for the reaction; however, this can be substituted for any other entry vector with requisite BsmBI cut sites. Once built, part plasmids can be assembled into transcriptional units.
Protocol source: NEB (https://www.neb.com/protocols/2020/01/15/golden-gate-assembly-protocol-for-using-neb-golden-gate-assembly-kit-bsmbi-v2-neb-e1602
) you can proceed to the assembly step of the part plasmid itself. The example reaction below shows pYTK001 used as the entry vector for the reaction; however, this can be substituted for any other entry vector with requisite BsmBI cut sites. Once built, part plasmids can be assembled into transcriptional units.
Protocol source: NEB (https://www.neb.com/protocols/2020/01/15/golden-gate-assembly-protocol-for-using-neb-golden-gate-assembly-kit-bsmbi-v2-neb-e1602 )
)
 2. Set up the following reaction mix:
3. Mix samples well by pipetting, then run the reaction on the thermocycler under the following conditions:
2. Set up the following reaction mix:
3. Mix samples well by pipetting, then run the reaction on the thermocycler under the following conditions:



