Back to Golden Gate Protocols
Part Plasmid Assembly
Once you have designed your part and either amplified with PCR or ordered the desired gBlock (as described
here) you can proceed to the assembly step of the part plasmid itself. The example reaction below shows pYTK001 used as the entry vector for the reaction; however, this can be substituted for any other entry vector with requisite BsmBI cut sites. Once built, part plasmids can be assembled into transcriptional units.
Protocol source: NEB (
https://www.neb.com/protocols/2020/01/15/golden-gate-assembly-protocol-for-using-neb-golden-gate-assembly-kit-bsmbi-v2-neb-e1602)
Assembly Reaction
Access the old non-kit golden gate assembly protocols here
1. Calculate the mass (in ng) required for 50 fmol of vector and 100 fmol of insert using NEB's NEBioCalculator:
https://nebiocalculator.neb.com/#!/dsdnaamt
2. Set up the following reaction mix:
- 50 fmol pYTK-001 plasmid = 82.68 ng [try to keep volume to 1-2μL]
- 100 fmol of insert DNA = 650 * insert length * 100x10^-6 = X ng [try to keep volume to less than 10 μL]
- 2 μL of NEB 10× T4 DNA ligase buffer
- 1 μL of NEB Golden Gate Enzyme Mix (BsmBI-v2)
- x μL water up to 20 μL total
3. Mix samples well by pipetting, then run the reaction on the thermocycler under the following conditions:
| Step |
Temperature |
Time |
| 1 |
42°C |
1.5 min |
| 2 |
16°C |
3 min |
| Cycles 1-2: |
Repeat 25x |
|
| 3 |
50°C |
5 min |
| 4 |
80°C |
10 min |
Optionally, you may have high enough efficiency if inserting one PCR product into the entry vector with NEB's short protocol with no cycling instead of the longer protocol:
| Step |
Temperature |
Time |
| 1 |
42°C |
5 min |
| 2 |
60°C |
5 min |
4. Transform 2 μL assembly reaction into
Electrocompetent or
Chemically Competent cells and plate on LB + Cam
Expected Results
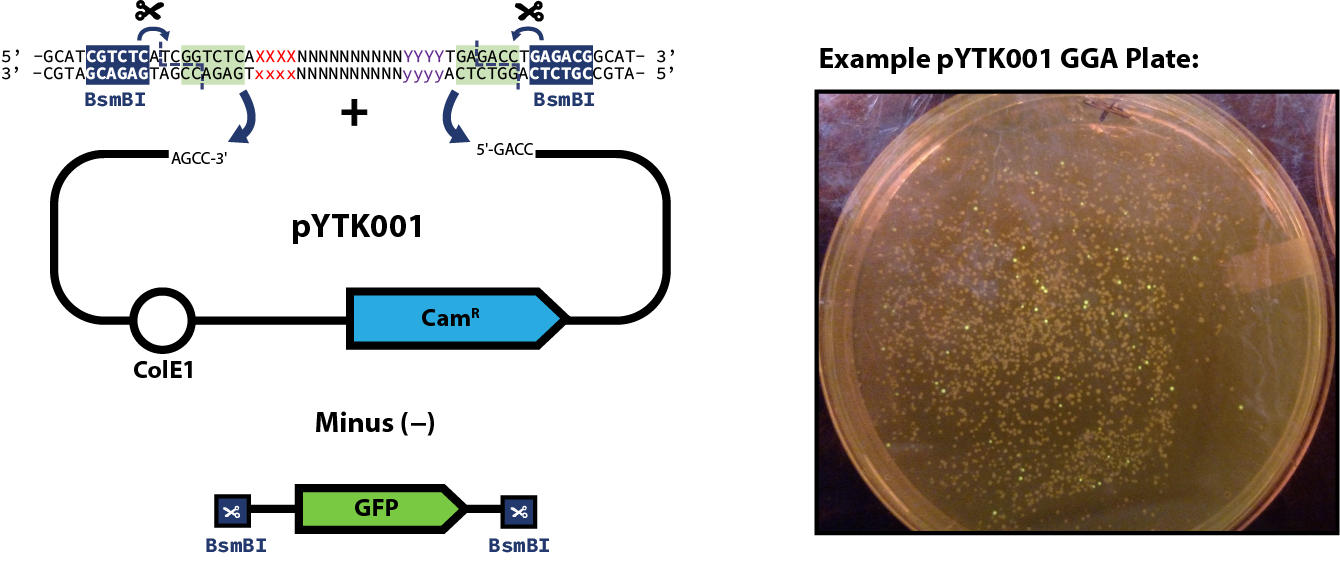
- (If using pYTK001 as your entry vector) Multiple non-fluorescent colonies
- Be sure to screen colonies for GFP expression on a blue light transilluminator, pick colonies that do not fluoresce!
- Cells containing properly assembled plasmids (confirmed through PCR and Sanger Sequencing)
If you are having repeated issues check out the Troubleshooting page!
Back to Golden Gate Protocols
Topic revision: r16 - 2022-06-30 - 20:57:43 - Main.JeffreyBarrick


Barrick Lab > BroadHostRangeToolkit > ProtocolsBTKMakeANewPartPlasmid

