
Difference: ProtocolsBTKMakeANewPartPlasmid (12 vs. 13)
Revision 132021-11-03 - PatrickLariviere
Part Plasmid AssemblyOnce you have designed your part and either amplified with PCR or ordered the desired gBlock (as described here | |||||||||||||
| Added: | |||||||||||||
| > > |
Protocol source: NEB (https://www.neb.com/protocols/2020/01/15/golden-gate-assembly-protocol-for-using-neb-golden-gate-assembly-kit-bsmbi-v2-neb-e1602 | ||||||||||||
Assembly ReactionAccess the old non-kit golden gate assembly protocols here | |||||||||||||
| Changed: | |||||||||||||
| < < | 1. Set up the following reaction mix: | ||||||||||||
| > > | 1. Calculate the mass (in ng) required for 50 fmol of vector and 100 fmol of insert using NEB's NEBioCalculator: https://nebiocalculator.neb.com/#!/dsdnaamt | ||||||||||||
| Added: | |||||||||||||
| > > | 2. Set up the following reaction mix: | ||||||||||||
| Changed: | |||||||||||||
| < < |
| ||||||||||||
| > > |
| ||||||||||||
| Deleted: | |||||||||||||
| < < |
| ||||||||||||
| |||||||||||||
| Changed: | |||||||||||||
| < < | 2. Mix samples well by pipetting, then run the reaction on the thermocycler under the following conditions:
| ||||||||||||
| > > | 3. Mix samples well by pipetting, then run the reaction on the thermocycler under the following conditions:
| ||||||||||||
| Deleted: | |||||||||||||
| < < |
| ||||||||||||
| Changed: | |||||||||||||
| < < | |||||||||||||
| > > | 4. Transform 2 μL assembly reaction into Electrocompetent or Chemically Competent cells and plate on LB + Cam | ||||||||||||
| Deleted: | |||||||||||||
| < < | 3. Transform 2 μL assembly reaction into Electrocompetent or Chemically Competent cells and plate on LB + Cam | ||||||||||||
Expected Results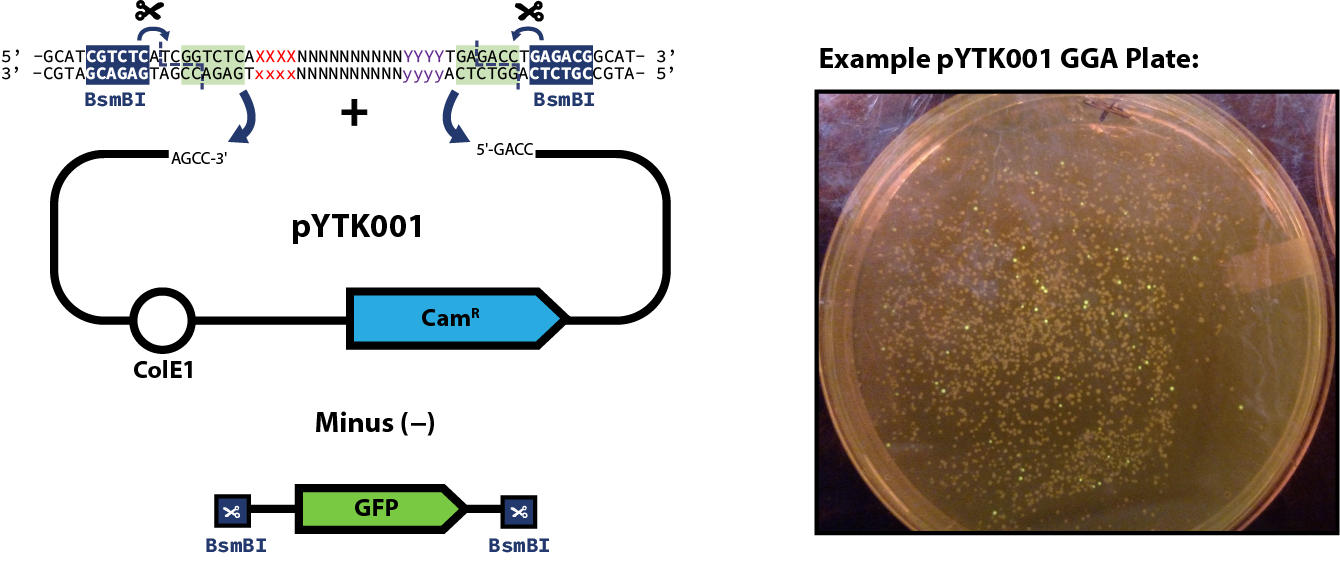
Back to Golden Gate Protocols
| |||||||||||||
View topic | History: r17 < r16 < r15 < r14 | More topic actions...
Ideas, requests, problems regarding TWiki? Send feedback