
Difference: ProtocolsCFUCounts (1 vs. 3)
Revision 32022-08-17 - KateElston
CFU CountsThis is an outline for a general protocol to assess the number of colony forming units in a sample using spot plating. This method cuts down on the number of plates and makes the process better for high throughput experiments. We have used this method to assess bacterial titer in insects and determine conjugation efficiency, but it has plenty of other practical uses.(Optional) Preparation of SampleThis first step is useful to prepare insect samples when calculating bacterial titer or when working with bacteria that are being transferred from one media condition to another (ie following conjugation when samples are transferred from nonselective DAP plates to selective plates). This preparation involves performing washes in a saline solution to remove media/surface contaminants. Insect samples:
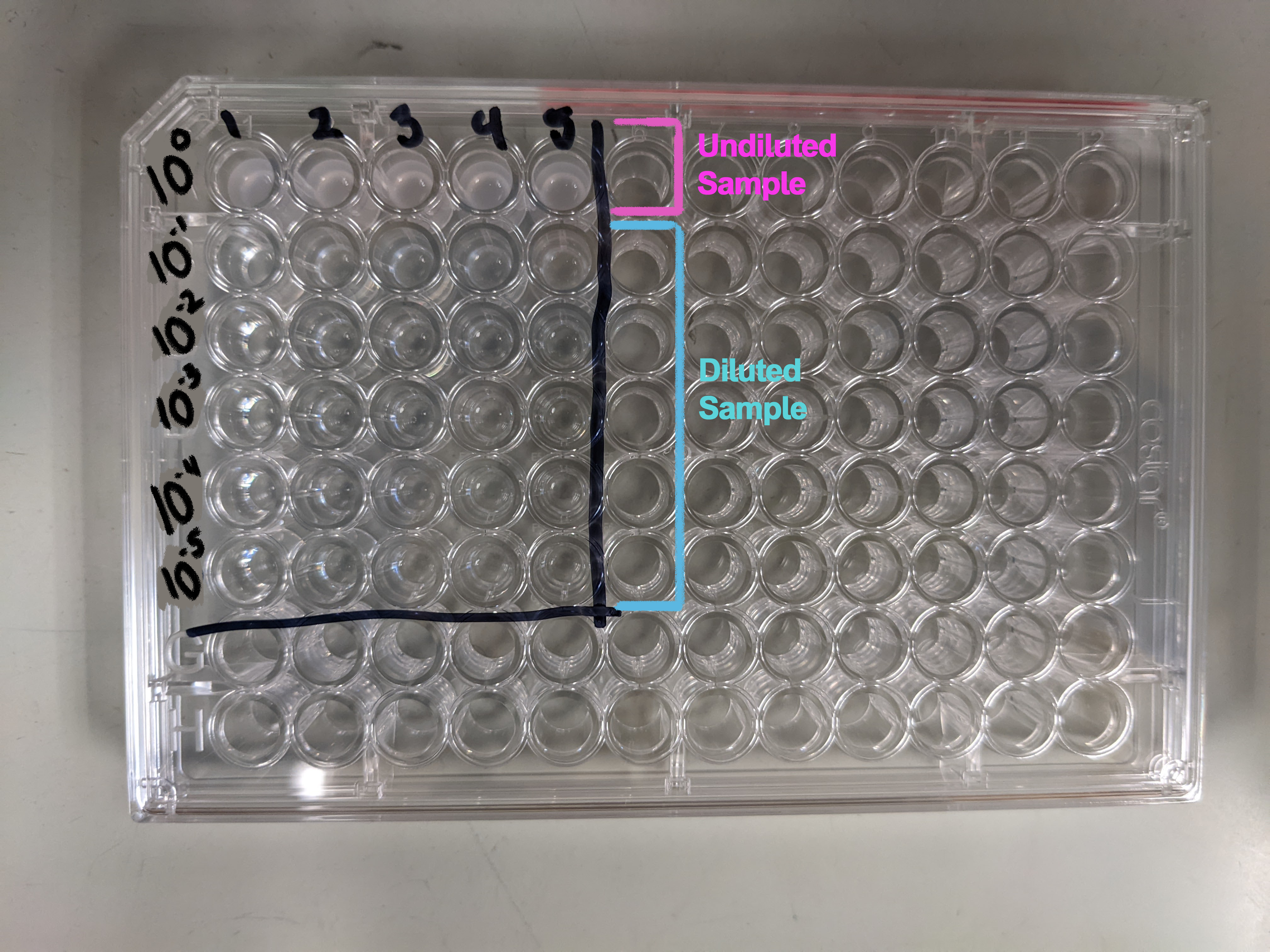
Sample Dilution and platingGenerally the dilutions you'll want to perform can be done in a 96-well plate. This makes the process compatible with multichannel pipets and the Opentron robot to speed up the process. The image below shows the plate setup with 1:10 dilutions ranging from 100 to 10-5. If, when performing this assay, you find that this range is not useful for your sample you can adjust it by changing the number or scale of the dilutions. What you need: Dilution:
| ||||||||
| Added: | ||||||||
| > > |  | |||||||
| ||||||||
| Changed: | ||||||||
| < < |
| |||||||
| > > |
| |||||||
CFU counting and calculations | ||||||||
| Added: | ||||||||
| > > |  | |||||||
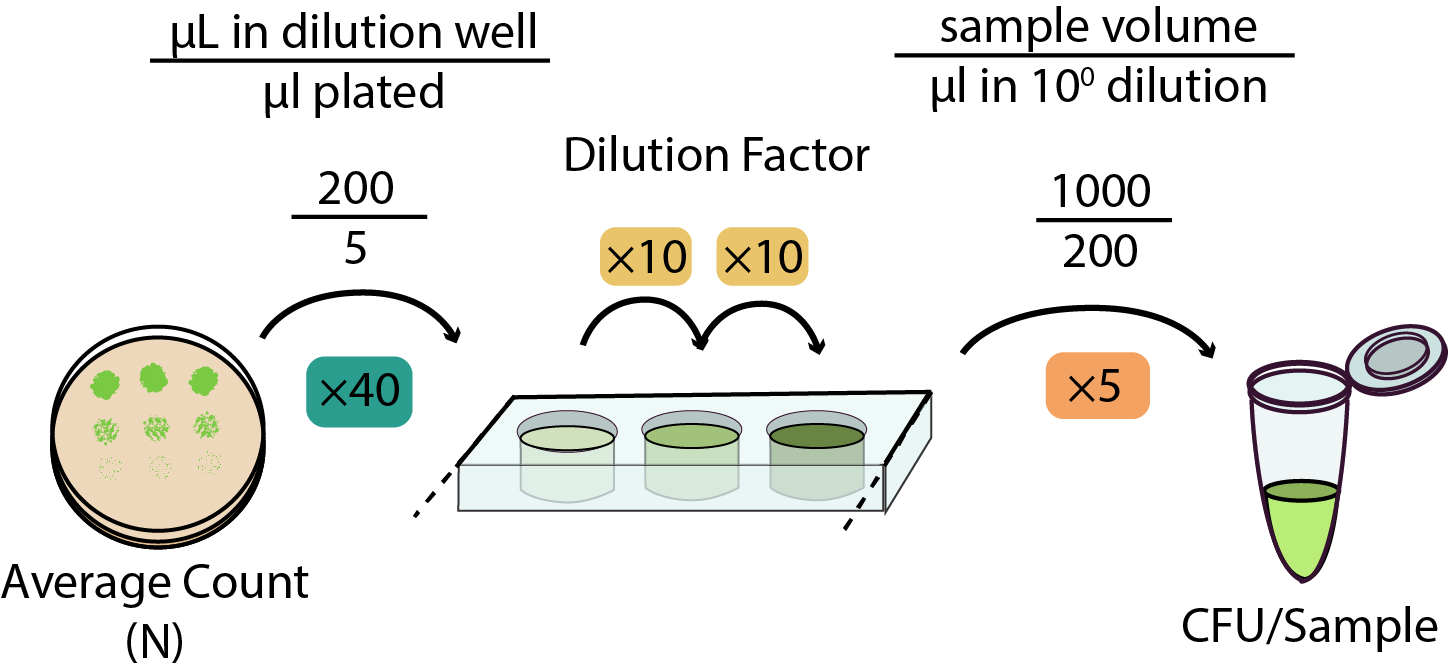
| To count your colonies it is important that they are visible and not running into each other! This can sometimes be challenging when working with 5 µL spots - you'll want to work out the appropriate amount of growth time for whichever bacteria you are testing in this assay. Counting recommendations: | ||||||||
| Changed: | ||||||||
| < < |
| |||||||
| > > |
| |||||||
Contributors to this protocol:--References:Elston et al., 2021 | ||||||||
| Added: | ||||||||
| > > | ||||||||
| Added: | ||||||||
| > > | ||||||||
| ||||||||
| Added: | ||||||||
| > > |
| |||||||
Revision 22022-08-05 - KateElston
CFU CountsThis is an outline for a general protocol to assess the number of colony forming units in a sample using spot plating. This method cuts down on the number of plates and makes the process better for high throughput experiments. We have used this method to assess bacterial titer in insects and determine conjugation efficiency, but it has plenty of other practical uses.(Optional) Preparation of SampleThis first step is useful to prepare insect samples when calculating bacterial titer or when working with bacteria that are being transferred from one media condition to another (ie following conjugation when samples are transferred from nonselective DAP plates to selective plates). This preparation involves performing washes in a saline solution to remove media/surface contaminants. Insect samples:
Sample Dilution and platingGenerally the dilutions you'll want to perform can be done in a 96-well plate. This makes the process compatible with multichannel pipets and the Opentron robot to speed up the process. The image below shows the plate setup with 1:10 dilutions ranging from 100 to 10-5. If, when performing this assay, you find that this range is not useful for your sample you can adjust it by changing the number or scale of the dilutions. What you need: Dilution:
CFU counting and calculationsTo count your colonies it is important that they are visible and not running into each other! This can sometimes be challenging when working with 5 µL spots - you'll want to work out the appropriate amount of growth time for whichever bacteria you are testing in this assay. Counting recommendations:
| ||||||||
| Changed: | ||||||||
| < < | To calculate the CFU/sample you can use this convenient spreadsheet to perform the calculation. However, to explain the process and enable you to make your own high throughput spreadsheets for many samples at a time please refer to the image below: | |||||||
| > > | To calculate the CFU/sample you can use this convenient spreadsheet to perform the calculation. However, to explain the process and enable you to make your own high-throughput calculation spreadsheet please refer to the image below which uses the values described in the protocol above as an example: | |||||||
| Added: | ||||||||
| > > |  | |||||||
| Added: | ||||||||
| > > | ||||||||
Contributors to this protocol:--References:Elston et al., 2021 | ||||||||
| Added: | ||||||||
| > > | ||||||||
| ||||||||
| Added: | ||||||||
| > > |
| |||||||
Revision 12022-08-05 - KateElston
CFU CountsThis is an outline for a general protocol to assess the number of colony forming units in a sample using spot plating. This method cuts down on the number of plates and makes the process better for high throughput experiments. We have used this method to assess bacterial titer in insects and determine conjugation efficiency, but it has plenty of other practical uses.(Optional) Preparation of SampleThis first step is useful to prepare insect samples when calculating bacterial titer or when working with bacteria that are being transferred from one media condition to another (ie following conjugation when samples are transferred from nonselective DAP plates to selective plates). This preparation involves performing washes in a saline solution to remove media/surface contaminants. Insect samples:
Sample Dilution and platingGenerally the dilutions you'll want to perform can be done in a 96-well plate. This makes the process compatible with multichannel pipets and the Opentron robot to speed up the process. The image below shows the plate setup with 1:10 dilutions ranging from 100 to 10-5. If, when performing this assay, you find that this range is not useful for your sample you can adjust it by changing the number or scale of the dilutions. What you need: Dilution:
CFU counting and calculationsTo count your colonies it is important that they are visible and not running into each other! This can sometimes be challenging when working with 5 µL spots - you'll want to work out the appropriate amount of growth time for whichever bacteria you are testing in this assay. Counting recommendations:
Contributors to this protocol:--References:Elston et al., 2021
|
View topic | History: r3 < r2 < r1 | More topic actions...
Ideas, requests, problems regarding TWiki? Send feedback