
Difference: GoldenGateAssemblyProtocolsMainPage (9 vs. 10)
Revision 102018-06-07 - DennisMishler
Golden Gate AssemblyThis page serves as the main repository for everything Golden Gate Assembly-related. This page links to a number of protocol pages that will help you design DNA sequences for Golden Gate Assembly, learn about the various different Golden Gate Assembly techniques and when we employ each one, and conduct the different types of Golden Gate Assembly reactions. Background & Design:Golden Gate Assembly (GGA) was first described in Engler C, Kandzia R, Marillonnet S (2008) and Engler C, Gruetzner R, Kandzia R, Marillonnet S (2009) as an efficient way to quickly assembly multiple DNA sequences, or parts, into a single plasmid. This molecular cloning method allows a researcher to simultaneously and directionally assemble multiple DNA fragments into a single piece using Type IIs restriction enzymes and T4/T7 DNA ligase. This assembly is performed in vitro. The most commonly used Type IIs enzymes are BsaI, BsmBI and BbsI. Unlike standard Type II restriction enzymes like EcoRI and BamHI, these enzymes cut DNA outside of their recognition sites and therefore can create non-palindromic overhangs. Since 256 potential overhang sequences are possible, multiple fragments of DNA can be assembled by using combinations of overhang sequences. Golden Gate Assembly Schematic: 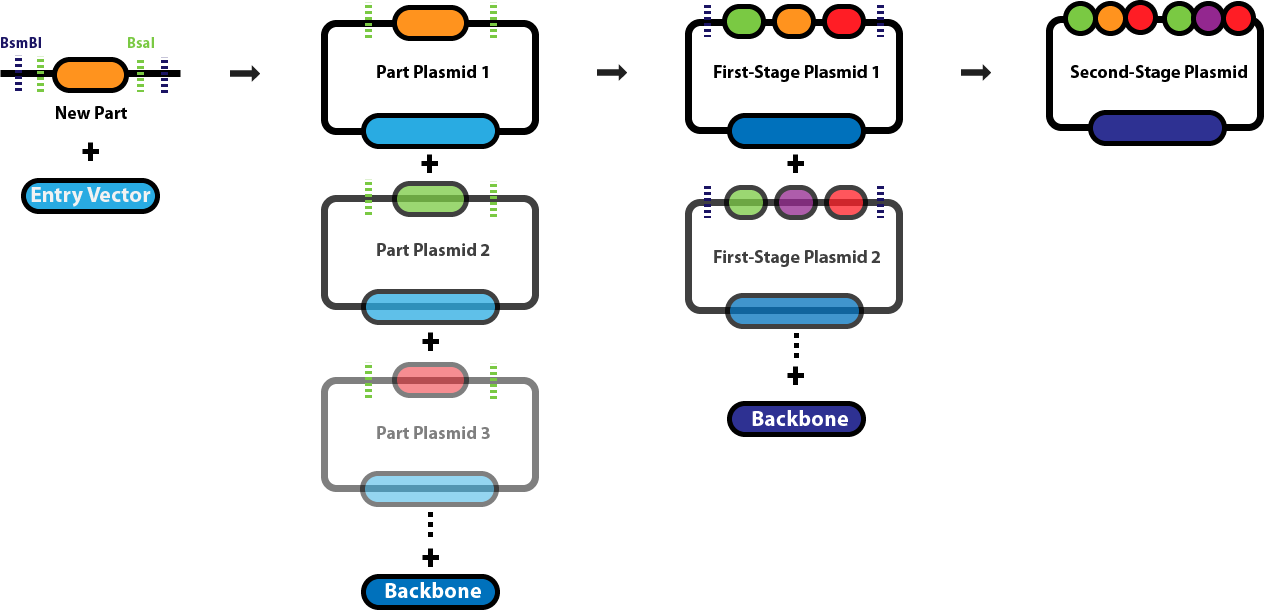
Golden Gate Assembly LinksGolden Gate Assembly - Start to Finish
References
Contributors
-- Main.KateElston - 29 Jan 2018 | |||||||||||||
| Added: | |||||||||||||
| > > |
The excel worksheet for calculating Golden Gate Assembly Reactions can be found in the attachment below. | ||||||||||||
| |||||||||||||
View topic | History: r16 < r15 < r14 < r13 | More topic actions...